
rsyncrosim: introduction to pipelines
Source:vignettes/a03_rsyncrosim_vignette_pipelines.Rmd
a03_rsyncrosim_vignette_pipelines.RmdThis vignette will cover how to implement model pipelines using the
rsyncrosim package within the
SyncroSim software
framework. For an overview of
SyncroSim and
rsyncrosim,
as well as a basic usage tutorial for rsyncrosim, see the
Introduction
to rsyncrosim vignette. To learn how to use iterations
in the rsyncrosim interface, see the
rsyncrosim:
introduction to uncertainty vignette.
SyncroSim Package: helloworldPipeline
To demonstrate how to link models in a pipeline using the
rsyncrosim interface, we will need the
helloworldPipeline
SyncroSim package. helloworldPipeline was designed to be a
simple package to introduce pipelines to SyncroSim modeling workflows.
Models (i.e. transformers) connected by pipelines allow the user to
implement multiple transformers in a modeling workflow and access
intermediate outputs of a transformer without having to create multiple
scenarios.
The package takes from the user 3 inputs, mMean, mSD, and b. For each iteration, a value m, representing the slope, is sampled from a normal distribution with mean of mMean and standard deviation of mSD. The b value represents the intercept. In the first model in the pipeline, these input values are run through a linear model, y=mt+b, where t is time, and the y value is returned as output. The second model takes y as input and calculates the cumulative sum of y over time, returning a new variable yCum as output.
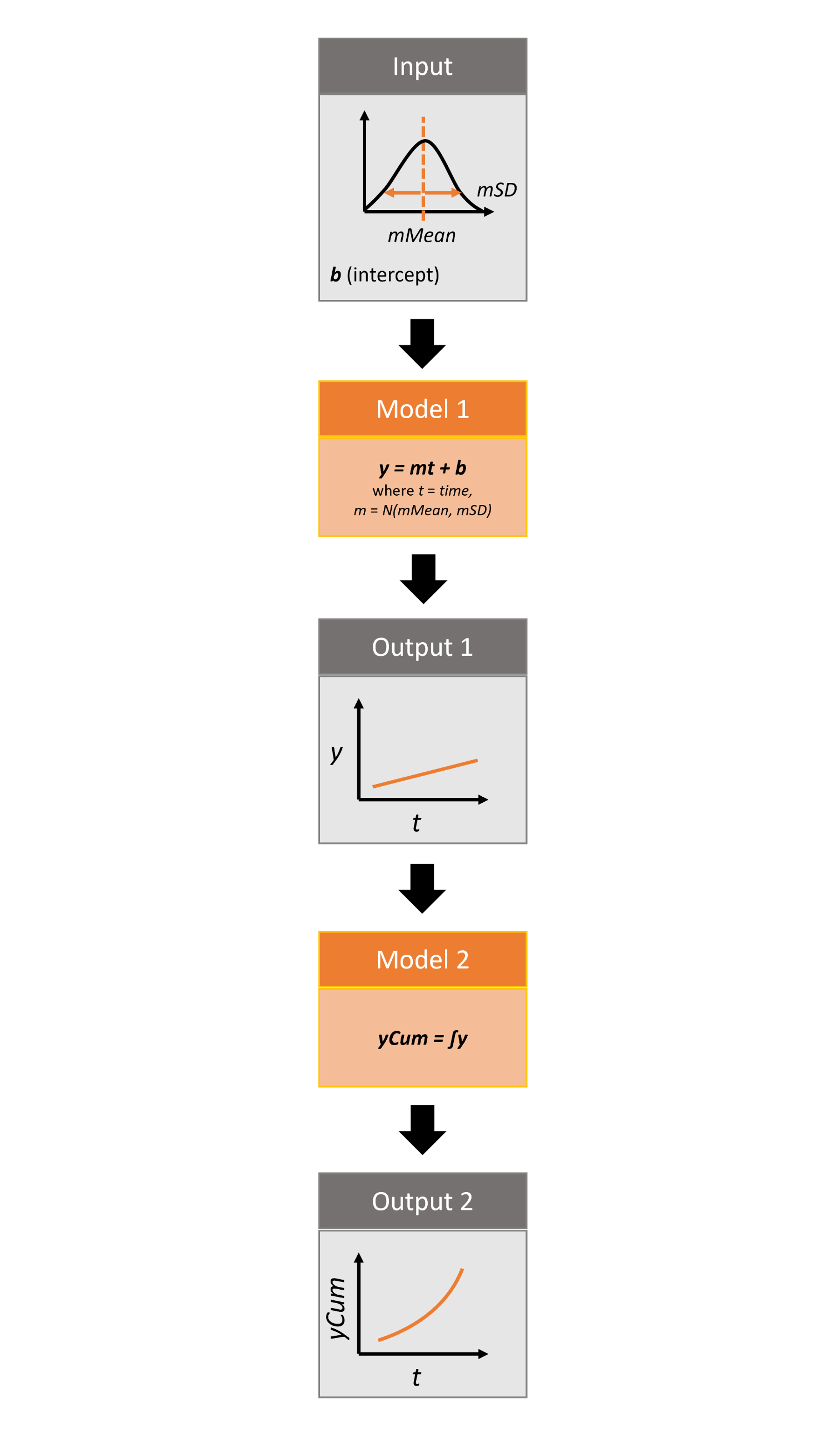
For more details on the different features of the
helloworldPipeline SyncroSim package, consult the SyncroSim
Enhancing
a Package: Linking Models tutorial.
Setup
Install SyncroSim
Before using rsyncrosim you will first need to
download and
install the SyncroSim software. Versions of SyncroSim exist for both
Windows and Linux.
Note: this tutorial was developed using
rsyncrosim version 2.0. To use rsyncrosim
version 2.0 or greater, SyncroSim version 3.0 or greater is
required.
Installing and loading R packages
You will need to install the rsyncrosim R package,
either using
CRAN or from
the rsyncrosim
GitHub
repository. Versions of rsyncrosim are available for
both Windows and Linux.
In a new R script, load the rsyncrosim package.
# Load R package for working with SyncroSim
library(rsyncrosim)Connecting R to SyncroSim using session()
Finish setting up the R environment for the rsyncrosim
workflow by creating a SyncroSim Session object. Use the
session() function to connect R to your installed copy of
the SyncroSim software.
mySession <- session("path/to/install_folder") # Create a Session based SyncroSim install folder
mySession <- session() # Using default install folder (Windows only)
mySession # Displays the Session object## class : Session
## filepath [character]: C:\PROGRA~1\SYNCRO~1
## silent [logical] : TRUE
## printCmd [logical] : FALSE
## condaFilepath [NULL]:Use the version() function to ensure you are using the
latest version of SyncroSim.
version(mySession)## [1] "3.1.24"Installing SyncroSim packages using
installPackage()
Install helloworldPipeline using the
rynscrosim function installPackage(). This
function takes a package name as input and then queries the SyncroSim
package server for the specified package.
# Install helloworldPipeline
installPackage("helloworldPipeline")## Package <helloworldPipeline v2.1.1> installedhelloworldPipeline should now be included in the package
list returned by the packages() function in
rsyncrosim:
# Get list of installed packages
packages()## name version
## 1 helloworldPipeline 2.1.1
## description
## 1 Example demonstrating how to use pipelines with an R model
## location
## 1 C:\\Users\\HannahAdams\\AppData\\Local\\SyncroSim\\Packages\\helloworldPipeline\\2.1.1
## status
## 1 OKCreate a modeling workflow
When creating a new modeling workflow from scratch, we need to create objects of the following scopes:
For more information on these scopes, see the Introduction
to rsyncrosim vignette.
Set up library, project, and scenario
# Create a new library
myLibrary <- ssimLibrary(name = "helloworldLibrary.ssim",
session = mySession,
packages = "helloworldPipeline",
overwrite = TRUE)## Package <helloworldPipeline v2.1.1> addedView model inputs using datasheet()
View the datasheets associated with your new scenario using the
datasheet() function from rsyncrosim.
# View all datasheets associated with a library, project, or scenario
datasheet(myScenario)## scope name displayName
## 26 scenario core_DistributionValue Distributions
## 27 scenario core_ExternalVariableValue External Variables
## 28 scenario core_Pipeline Pipeline
## 29 scenario core_SpatialMultiprocessing Spatial Multiprocessing
## 30 scenario helloworldPipeline_InputDatasheet Inputs
## 31 scenario helloworldPipeline_IntermediateDatasheet Intermediate Outputs
## 32 scenario helloworldPipeline_OutputDatasheet Outputs
## 33 scenario helloworldPipeline_RunControl Run ControlFrom the list of datasheets above, we can see that there are four
datasheets specific to the helloworldPipeline package,
including an Inputs datasheet, an
Intermediate Outputs datasheet, and an Outputs
datasheet. These three datasheets are connected by transformers. The
values from the Inputs datasheet are used as the input for
the first transformer, which transforms the input data to
output data through a series of model calculations. The output data from
the first transformer is contained within the
Intermediate Outputs datasheet. The values from the
Intermediate Outputs datasheet are then used as input for
the second transformer. The output from the second transformer is stored
in the Outputs datasheet.
Configure model inputs using datasheet() and
addRow()
Currently our input scenario datasheets are empty! We need to add
some values to our Inputs datasheet
(InputDatasheet) and Run Control datasheet
(RunControl) so we can run our model. We also need to add
some information to the core Pipeline datasheet to specify
which transformers are run in which order.
Inputs Datasheet
First, assign the contents of the Inputs datasheet to a
new data frame variable using datasheet(), then check the
columns that need input values.
# Load Inputs datasheet to a new R data frame
myInputDataframe <- datasheet(myScenario,
name = "helloworldPipeline_InputDatasheet")
# Check the columns of the input data frame
str(myInputDataframe)## 'data.frame': 0 obs. of 3 variables:
## $ mMean: num
## $ mSD : num
## $ b : numThe Inputs datasheet requires three values:
-
mMean: the mean of the slope normal distribution. -
mSD: the standard deviation of the slope normal distribution. -
b: the intercept of the linear equation.
Add these values to a new data frame, then use the
addRow() function from rsyncrosim to update
the input data frame
# Create input data and add it to the input data frame
myInputRow <- data.frame(mMean = 2, mSD = 4, b = 3)
myInputDataframe <- addRow(myInputDataframe, myInputRow)
# Check values
myInputDataframe## mMean mSD b
## 1 2 4 3Finally, save the updated R data frame to a SyncroSim datasheet using
saveDatasheet().
# Save input R data frame to a SyncroSim datasheet
saveDatasheet(ssimObject = myScenario,
data = myInputDataframe,
name = "helloworldPipeline_InputDatasheet")## Datasheet <helloworldPipeline_InputDatasheet> savedRun Control Datasheet
The Run Control datasheet provides information about how
many time steps and iterations to use in the model. Here, we set the
number of iterations, as well as the minimum and maximum time steps for
our model. Let’s take a look at the columns that need input values.
# Load Run Control datasheet to a new R data frame
runSettings <- datasheet(myScenario, name = "helloworldPipeline_RunControl")
# Check the columns of the Run Control data frame
str(runSettings)## 'data.frame': 0 obs. of 3 variables:
## $ MinimumTimestep : num
## $ MaximumTimestep : num
## $ MaximumIteration: numThe Run Control datasheet requires the following 3 columns:
-
MaximumIteration: total number of iterations to run the model for. -
MinimumTimestep: the starting time point of the simulation. -
MaximumTimestep: the end time point of the simulation.
Note: A fourth hidden column, MinimumIteration,
also exists in the Run Control datasheet (default=1).
We’ll add this information to a new data frame and then add it to the
Run Control data frame using addRow().
# Create Run Control data and add it to the Run Control data frame
runSettingsRow <- data.frame(MaximumIteration = 5,
MinimumTimestep = 1,
MaximumTimestep = 10)
runSettings <- addRow(runSettings, runSettingsRow)
# Check values
runSettings## MinimumTimestep MaximumTimestep MaximumIteration
## 1 1 10 5Finally, save the R data frame to a SyncroSim datasheet using
saveDatasheet().
# Save Run Control R data frame to a SyncroSim datasheet
saveDatasheet(ssimObject = myScenario, data = runSettings,
name = "helloworldPipeline_RunControl")## Datasheet <helloworldPipeline_RunControl> savedPipeline Datasheet
To implement pipelines in our package, we need to specify the order
in which to run the transformers in our pipeline by adding data to the
Pipeline datasheet. The Pipeline datasheet is
a built-in SyncroSim datasheet, meaning that it comes with every
SyncroSim library regardless of which packages that library uses. We
access it using the “core_” prefix with the datasheet()
function.
From viewing the structure of the Pipeline datasheet we
know that the StageNameId is a factor with two levels:
- Hello World Pipeline 1 (R)
- Hello World Pipeline 2 (R)
We will set the data for this datasheet such that
Hello World Pipeline 1 (R) is run first, then
Hello World Pipeline 2 (R). This way, the output from
Hello World Pipeline 1 (R) is used as the input for
Hello World Pipeline 2 (R).
# Load Pipeline datasheet to a new R data frame
myPipelineDataframe <- datasheet(myScenario, name = "core_Pipeline")
# Check the columns of the Pipeline data frame
str(myPipelineDataframe)## 'data.frame': 0 obs. of 2 variables:
## $ StageNameId: Factor w/ 2 levels "Hello World Pipeline 1 (R)",..:
## $ RunOrder : num
# Create Pipeline data and add it to the Pipeline data frame
myPipelineRow <- data.frame(StageNameId = c("Hello World Pipeline 1 (R)",
"Hello World Pipeline 2 (R)"),
RunOrder = c(1, 2))
myPipelineDataframe <- addRow(myPipelineDataframe, myPipelineRow)
# Check values
myPipelineDataframe## StageNameId RunOrder
## 1 Hello World Pipeline 1 (R) 1
## 2 Hello World Pipeline 2 (R) 2
# Save Pipeline R data frame to a SyncroSim datasheet
saveDatasheet(ssimObject = myScenario, data = myPipelineDataframe,
name = "core_Pipeline")## Datasheet <core_Pipeline> savedRun Scenarios
Setting the number of multiprocessing jobs
If we have a large model and we want to parallelize the run using multiprocessing, we can modify the library-scoped “core_Multiprocessing” datasheet. Since we are using five iterations in our model, we will set the number of jobs to five so each multiprocessing core will run a single iteration.
# Load list of available library-scoped datasheets
datasheet(myLibrary)## scope name displayName
## 1 library core_Backup Backup
## 2 library core_JlConfig Julia
## 3 library core_Multiprocessing Multiprocessing
## 4 library core_Option Options
## 5 library core_ProcessorGroupOption Processor Group Options
## 6 library core_ProcessorGroupValue Processor Group Values
## 7 library core_PublishDatasheet PublishDatasheet
## 8 library core_PyConfig Python
## 9 library core_RConfig R
## 10 library core_Setting Settings
## 11 library core_SpatialMultiprocessingOption Spatial Multiprocessing Option
## 12 library core_SpatialOption Spatial Options
## 13 library core_SysFolder Folders
## 14 library core_Terminology Terminology
# Load the library-scoped multiprocessing datasheet
multiprocess <- datasheet(myLibrary, name = "core_Multiprocessing")## [1] "Note: MaximumJobs should be between 1 and 9999"
# Check required inputs
str(multiprocess)## 'data.frame': 1 obs. of 4 variables:
## $ EnableMultiprocessing : logi FALSE
## $ MaximumJobs : num 15
## $ EnableMultiScenario : logi FALSE
## $ EnableCopyExternalFiles: logi NA
# Enable multiprocessing
multiprocess$EnableMultiprocessing <- TRUE
# Set maximum number of jobs to 5
multiprocess$MaximumJobs <- 4
# Save multiprocessing configuration
saveDatasheet(ssimObject = myLibrary,
data = multiprocess,
name = "core_Multiprocessing")## Datasheet <core_Multiprocessing> savedSetting run parameters with run()
Now, when we run our scenario, it will use the desired multiprocessing configuration.
# Run the first scenario we created
myResultScenario <- run(myScenario)## [1] "Running scenario [1] My first scenario"## This model uses Conda environments, but no Conda installation was found. Using local environment.Once the run is complete, we can compare the original scenario to the
result scenario to see which datasheets have been modified. Using the
datasheet() function with the optional
argument set to TRUE, we see that data has been added to
both the Intermediate Outputs and Outputs
datasheets after running the scenario (see data column
below).
# Datasheets for original scenario
datasheet(myScenario, optional = TRUE)## scope package name
## 5 scenario core core_DistributionValue
## 7 scenario core core_ExternalVariableValue
## 15 scenario core core_Pipeline
## 22 scenario core core_SpatialMultiprocessing
## 30 scenario helloworldPipeline helloworldPipeline_InputDatasheet
## 31 scenario helloworldPipeline helloworldPipeline_IntermediateDatasheet
## 32 scenario helloworldPipeline helloworldPipeline_OutputDatasheet
## 33 scenario helloworldPipeline helloworldPipeline_RunControl
## displayName isSingle displayMember data scenario
## 5 Distributions FALSE N/A FALSE 1
## 7 External Variables FALSE N/A FALSE 1
## 15 Pipeline FALSE N/A TRUE 1
## 22 Spatial Multiprocessing TRUE N/A FALSE 1
## 30 Inputs TRUE N/A TRUE 1
## 31 Intermediate Outputs FALSE N/A FALSE 1
## 32 Outputs FALSE N/A FALSE 1
## 33 Run Control TRUE N/A TRUE 1
# Datasheets for result scenario
datasheet(myResultScenario, optional = TRUE)## scope package name
## 5 scenario core core_DistributionValue
## 7 scenario core core_ExternalVariableValue
## 15 scenario core core_Pipeline
## 22 scenario core core_SpatialMultiprocessing
## 30 scenario helloworldPipeline helloworldPipeline_InputDatasheet
## 31 scenario helloworldPipeline helloworldPipeline_IntermediateDatasheet
## 32 scenario helloworldPipeline helloworldPipeline_OutputDatasheet
## 33 scenario helloworldPipeline helloworldPipeline_RunControl
## displayName isSingle displayMember data scenario
## 5 Distributions FALSE N/A FALSE 1
## 7 External Variables FALSE N/A FALSE 1
## 15 Pipeline FALSE N/A TRUE 1
## 22 Spatial Multiprocessing TRUE N/A FALSE 1
## 30 Inputs TRUE N/A TRUE 1
## 31 Intermediate Outputs FALSE N/A TRUE 1
## 32 Outputs FALSE N/A TRUE 1
## 33 Run Control TRUE N/A TRUE 1View results
The next step is to view the output datasheets added to the result scenario when it was run.
Viewing intermediate results with datasheet()
First, we will view the Intermediate Outputs datasheet
from the result scenario. We can load the result tables using the
datasheet() function. The Intermediate Outputs
datasheet corresponds to the results from the
Hello World Pipeline 1 transformer stage.
# Results of first scenario
resultsSummary <- datasheet(myResultScenario,
name = "helloworldPipeline_IntermediateDatasheet")
# View results table
head(resultsSummary)## Iteration Timestep y
## 1 1 1 11.86268
## 2 1 2 20.72536
## 3 1 3 29.58804
## 4 1 4 38.45073
## 5 1 5 47.31341
## 6 1 6 56.17609We can see that for every timestep in an iteration we have a new value of y corresponding to y=mt+b.
Viewing final results with datasheet()
Now, we will view the final output datasheet from the result
scenario. Again, we will use datasheet() to load the result
table. The Outputs datasheet corresponds to the results
from the Hello World Pipeline 2 transformer stage.
# Results of first scenario
resultsSummary <- datasheet(myResultScenario,
name = "helloworldPipeline_OutputDatasheet")
# View results table
head(resultsSummary)## Iteration Timestep yCum
## 1 1 1 11.86268
## 2 1 2 32.58804
## 3 1 3 62.17609
## 4 1 4 100.62682
## 5 1 5 147.94022
## 6 1 6 204.11631We can see for each timestep in an iteration, we have a new value of yCum, representing the cumulative value of y over time.